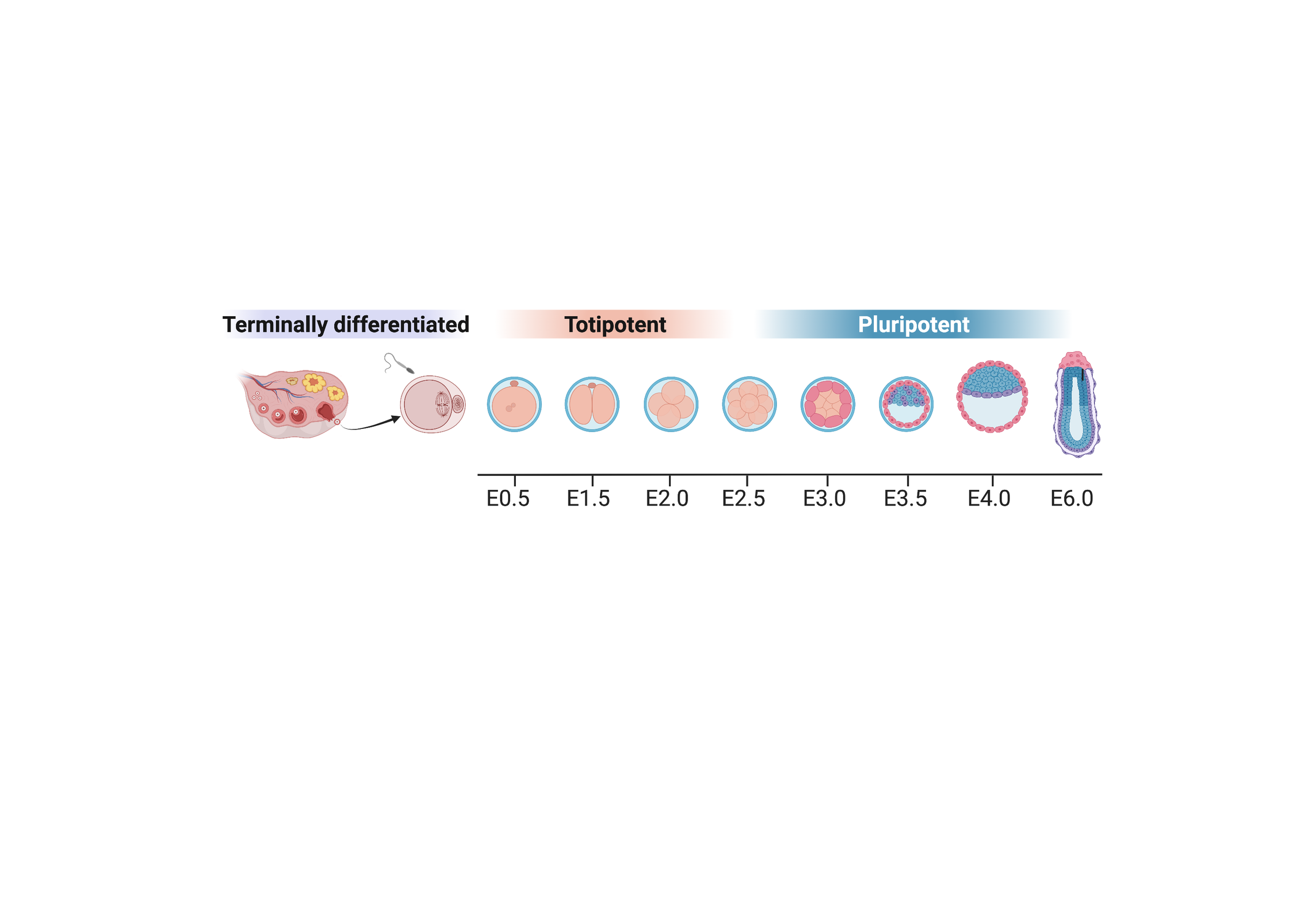
A new life begins at fertilization of the egg by the sperm. Following fertilization, the terminally differentiated gametes are converted into a totipotent zygote. During parental-to-zygotic transition, much of the parental epigenomes are reset except for imprinted loci. This resetting process is known as epigenetic reprogramming. Our lab aims to understand epigenetic reprogramming by studying 1) chromatin dynamics and functions during parental-to-zygotic transition, 2) key factors required for totipotency acquisition, and 3) biological significance of transposon expression dynamics in early development. In addition, we also focus on mechanisms underlying lineage- and cell type-specific genomic imprinting.
We implement interdisciplinary approaches to address these fundamental questions. The approaches include embryo micro-manipulation, stem cell biology, (epi)genome editing, ultra-low input epigenome profiling, rapid protein degradation, high throughput genome-wide sequencing, and computational techniques.
Research area #1
Chromatin dynamics and functions in early development Recent advances in ultra-low input techniques have enabled genome-wide profiling of chromatin modifications in oocytes and early embryos, which have been challenging to study due to scarce materials. Given their highly dynamic nature, oocytes and early embryos are becoming unique in vivo model systems for studying establishment, maintenance, and erasure of chromatin features. For example, our recent work on H3K27me3 and H2AK119ub1 reprogramming dynamics in mouse early embryos has provided unique insights regarding Polycomb gene regulations. Importantly, as early developmental window covers critical molecular events such as zygotic genome activation and totipotency acquisition, understanding chromatin dynamics during these processes is of great interest in the fields of reproductive and developmental biology.
Research area #2:
Functions of transposable elements (TEs) in early development About 50% of human genome are made up of TEs, which are also known as “jumping genes”. Given their threats to genome integrity, TEs are typically silenced by the host. Over evolutionary timescales, TEs accumulate mutations and majority of them have become immobilized. However, TEs often retain their regulatory sequences and can be repurposed to control host gene expression. TEs have been reported to function as promoters and enhancers and to modulate 3D chromatin. However, it remains unknown to what extent TEs impact genome function and regulate physiological processes. Remarkably, certain TEs are dynamically reactivated in early embryos, which coincides with epigenetic reprogramming and totipotency. Our lab aims to systemically perturb TEs to reveal their roles in embryonic development.
Research area #3:
Mechanisms and functions of placental imprinting Genomic imprinting is an epigenetic phenomenon that genes are transcribed in a parental allele-specific manner. Genomic imprinting is essential for mammalian development. Placenta has the most imprinted genes, highlighting the importance of genomic imprinting for this organ. There are two major imprinting mechanisms. The canonical imprinting is primarily regulated by germ-line differential DNA methylation, whereas recently discovered non-canonical imprinting is mainly controlled by oocyte-derived H3K27me3. Both canonical DNA methylation imprinting and non-canonical H3K27me3 imprinting are critical for placental function. Our future work will focus on how placental imprinting regulates nutrient acquisition and fetal growth, which will have important implications for pregnancy-related pathologies.